-Search query
-Search result
Showing all 49 items for (author: binshtein & e)

EMDB-41505: 
Hemagglutinin-neuraminidase from Human parainfluenza virus type 3: complex with rPIV3-23 and rPIV3-28 Fabs
Method: single particle / : Otrelo-Cardoso AR, Jardetzky TS

EMDB-41506: 
Human parainfluenza virus type 3 prefusion F trimer in complex with rPIV3-18 Fab
Method: single particle / : Otrelo-Cardoso AR, Jardetzky TS

EMDB-28199: 
Cryo-EM map of SARS-CoV-2 Omicron BA.2 spike in complex with 2130-1-0114-112
Method: single particle / : Binshtein E, Crowe JE

PDB-8ekd: 
Cryo-EM map of SARS-CoV-2 Omicron BA.2 spike in complex with 2130-1-0114-112
Method: single particle / : Binshtein E, Crowe JE
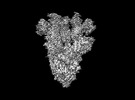
EMDB-28198: 
Cryo-EM map of SARS-CoV-2 Omicron BA.2 spike in complex with LLNL-199
Method: single particle / : Binshtein E, Crowe JE

EMDB-41075: 
SARS-CoV-2 spike in complex with Fab 71281-33
Method: single particle / : Binshtein E, Crowe JE

EMDB-41076: 
SARS-CoV-2 spike in complex with Fab 71281-33 (2)
Method: single particle / : Binshtein E, Crowe JE

EMDB-41503: 
XptA2 wild type
Method: single particle / : Martin CL, Binshtein EM, Aller SG

PDB-8tqe: 
XptA2 wild type
Method: single particle / : Martin CL, Binshtein EM, Aller SG

EMDB-26735: 
Hantavirus ANDV Gn(H) protein in complex with 2 Fabs ANDV-5 and ANDV-34
Method: single particle / : Binshtein E, Crowe JE
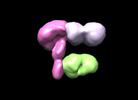
EMDB-26736: 
Hantavirus MAPV Gn(H)/Gc protein in complex with 2 Fabs SNV-24 and SNV-53
Method: single particle / : Binshtein E, Crowe JE

EMDB-27318: 
CryoEM structure of Hantavirus ANDV Gn(H) protein complex with 2Fabs ANDV-5 and ANDV-34
Method: single particle / : Binshtein E, Crowe JE

PDB-8dbz: 
CryoEM structure of Hantavirus ANDV Gn(H) protein complex with 2Fabs ANDV-5 and ANDV-34
Method: single particle / : Binshtein E, Crowe JE

EMDB-25102: 
CryoEM structure of Venezuelan Equine Encephalitis virus (VEEV) TC-83 strain VLP
Method: single particle / : Binshtein E, Crowe JE

EMDB-25103: 
CryoEM structure of Venezuelan Equine Encephalitis virus (VEEV) TC-83 strain VLP in complex with Fab hVEEV-63
Method: single particle / : Binshtein E, Crowe JE

EMDB-25104: 
CryoEM structure of Venezuelan Equine Encephalitis virus (VEEV) TC-83 strain VLP in complex with Fab hVEEV-63 (focus refine of the asymmetric unit)
Method: single particle / : Binshtein E, Crowe JE

PDB-7sfu: 
CryoEM structure of Venezuelan Equine Encephalitis virus (VEEV) TC-83 strain VLP
Method: single particle / : Binshtein E, Crowe JE

PDB-7sfv: 
CryoEM structure of Venezuelan Equine Encephalitis virus (VEEV) TC-83 strain VLP in complex with Fab hVEEV-63
Method: single particle / : Binshtein E, Crowe JE

PDB-7sfw: 
CryoEM structure of Venezuelan Equine Encephalitis virus (VEEV) TC-83 strain VLP in complex with Fab hVEEV-63 (focus refine of the asymmetric unit)
Method: single particle / : Binshtein E, Crowe JE

EMDB-23488: 
Hendra virus receptor binding protein in complex with HENV-103 and HENV-117 Fabs
Method: single particle / : Binshtein E, Crowe JE

EMDB-23154: 
Negative stain EM map of SARS-CoV-2 spike protein (trimer) with Fab COV2-2489 (NTD)
Method: single particle / : Binshtein E, Crowe JE

EMDB-23155: 
Negative stain EM map of SARS-CoV-2 spike protein (trimer) with Fab COV2-2676 (NTD)
Method: single particle / : Binshtein E, Crowe JE

EMDB-22188: 
CryoEM structure of Chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-143 Antibody
Method: single particle / : Williamson LE, Gilliland T, Yadav PK, Binshtein E, Bombardi R, Kose N, Nargi RS, Sutton RM, Armstrong E, Carnahan RH, Walker LM, Kim AS, Fox J, Diamond MS, Ohi M, Klimstra WB, Crowe JE

EMDB-22223: 
CryoEM structure of Chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-33 Antibody
Method: single particle / : Williamson LE, Gilliland T, Yadav PK, Binshtein E, Bombardi R, Kose N, Nargi RS, Sutton RM, Armstrong E, Carnahan RH, Walker LM, Kim AS, Fox J, Diamond MS, Ohi M, Klimstra WB, Crowe JE

EMDB-22276: 
CryoEM structure of Eastern Equine Encephalitis (EEEV) VLP
Method: single particle / : Binshtein E, Crowe JE

EMDB-22277: 
CryoEM structure of Eastern Equine Encephalitis (EEEV) VLP with Fab EEEV-143.
Method: single particle / : Binshtein E, Crowe JE

PDB-6xo4: 
CryoEM structure of Eastern Equine Encephalitis (EEEV) VLP
Method: single particle / : Binshtein E, Crowe JE

PDB-6xob: 
CryoEM structure of Eastern Equine Encephalitis (EEEV) VLP with Fab EEEV-143.
Method: single particle / : Binshtein E, Crowe JE

EMDB-22627: 
Negative stain EM map of SARS-COV-2 spike protein (trimer) with Fab COV2-2082
Method: single particle / : Binshtein E, Crowe JE

EMDB-22628: 
Negative stain EM map of SARS-COV-2 spike protein (trimer) with Fab COV2-2479
Method: single particle / : Binshtein E, Crowe JE

EMDB-22148: 
Negative stain EM map of SARS-COV-2 spike protein open RBD (trimer) with Fab COV2-2096
Method: single particle / : Binshtein E, Crowe JE

EMDB-22149: 
Negative stain EM map of SARS-COV-2 spike protein (trimer) with Fab COV2-2832
Method: single particle / : Binshtein E, Crowe JE

EMDB-22147: 
Negative stain EM map of SARS-COV-2 spike protein closed RBD (trimer) with Fab COV2-2096
Method: single particle / : Binshtein E, Crowe JE

EMDB-21965: 
Negative stain EM map of SARS CoV-2 spike protein (trimer)
Method: single particle / : Binshtein E

EMDB-21974: 
Negative stain EM map of SARS-CoV-2 spike protein (trimer) with Fab COV2-2165
Method: single particle / : Binshtein E

EMDB-21975: 
Negative stain EM map of SARS-CoV-2 spike protein (trimer) with Fab COV2-2196
Method: single particle / : Binshtein E

EMDB-21976: 
Negative stain EM map of SARS-CoV-2 spike protein (trimer) with Fab COV2-2130
Method: single particle / : Binshtein E

EMDB-21977: 
Negative stain EM map of SARS-CoV-2 spike protein (trimer) with Fab COV2-2130 and Fab COV2-2196
Method: single particle / : Binshtein E

EMDB-21101: 
Mouse retromer (VPS26/VPS35/VPS29) tetramer of heterotrimers
Method: single particle / : Kendall AK, Jackson LP

EMDB-21116: 
Mouse retromer (VPS26/VPS35/VPS29) chain interface I (VPS35/VPS35 dimer)
Method: single particle / : Kendall AK, Jackson LP

EMDB-21117: 
Mouse retromer (VPS26/VPS35/VPS29) dimer of heterotrimers
Method: single particle / : Kendall AK, Jackson LP

EMDB-21118: 
Mouse retromer (VPS26/VPS35/VPS29) chain interface II (VPS26/VPS26 dimer)
Method: single particle / : Kendall AK, Jackson LP

EMDB-21119: 
Mouse retromer sub-structure: VPS35/VPS35 curved dimer
Method: single particle / : Kendall AK, Jackson LP

EMDB-21135: 
Mouse retromer sub-structure: VPS35/VPS35 flat dimer
Method: single particle / : Kendall AK, Jackson LP

EMDB-21136: 
Mouse retromer (VPS26/VPS35/VPS29) heterotrimer
Method: single particle / : Kendall AK, Jackson LP

PDB-6vab: 
Mouse retromer sub-structure: VPS35/VPS35 flat dimer
Method: single particle / : Kendall AK, Jackson LP

PDB-6vac: 
Mouse retromer (VPS26/VPS35/VPS29) heterotrimer
Method: single particle / : Kendall AK, Jackson LP
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search





 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
